Similar Genome Finder Service¶
Overview¶
The Similar Genome Finder Service will find similar public genomes in PATRIC or compute genome distance estimation using Mash/MinHash. It returns a set of genomes matching the specified similarity criteria.
Using the Similar Genome Finder Service¶
The Similar Genome Finder submenu option under the Services main menu (Genomics category) opens the Similar Genome Finder input form (shown below).
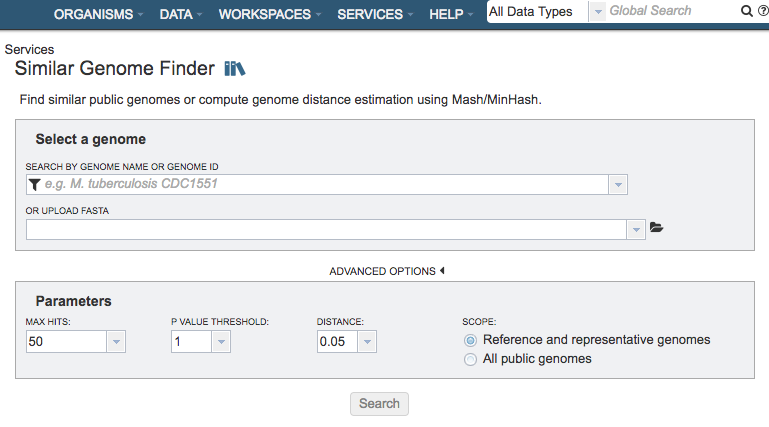
Select a Genome¶
Specifies the genome to use as the basis for finding other similar genomes
Search by Genome Name or Genome ID¶
Selection box for specifying genome in PATRIC to use as the basis of comparison
Or Upload FASTA¶
Alternate option for uploading a FASTA file to use as the basis of comparison. Note: You must be logged into PATRIC to use this option.
Advanced Options¶
Parameters¶
Max Hits: The maximum number of matching genomes to return.
P-Value Threshold: Sets the maximum allowable p-value associated with the Mash Jaccard estimate used in calculating the distance.
Distance: Mash distance, which estimates the rate of sequence mutation under as simple evolutionary model using k-mers. The Distance parameter sets the maximum Mash distance to include in the Similar Genome Finder Service results. Mash distances are probabilistic estimates associated with p-values.
Scope: Option for limiting the search to only Reference and Representative genomes, or all genomes in PATRIC.
Buttons¶
Search: Launches the similar genome finder job.
Output Results¶
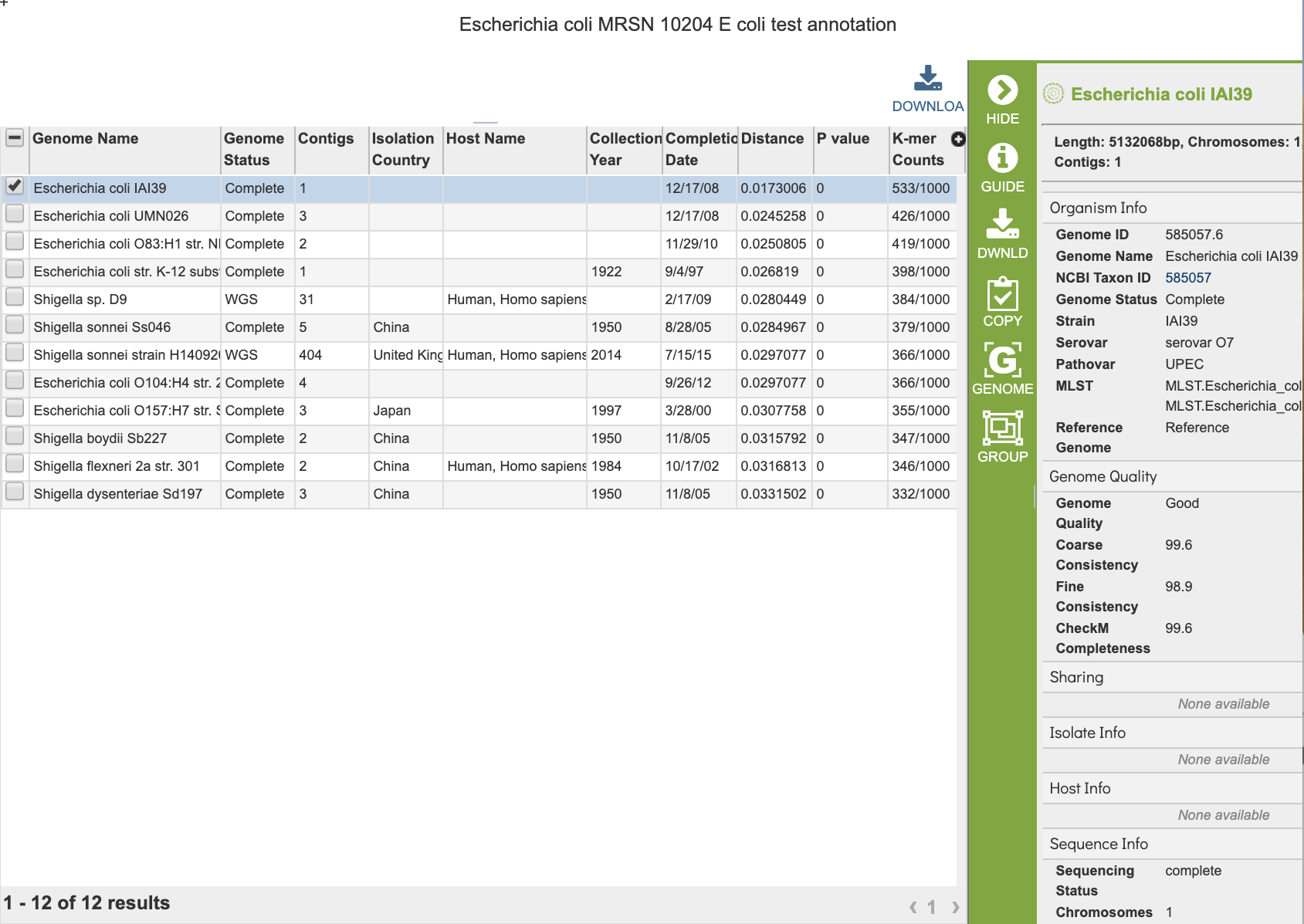
The Similar Genome Finder Service generates a table of matching genomes based on the options chosen.
Action buttons¶
After selecting one of the output files by clicking it, a set of options becomes available in the vertical green Action Bar on the right side of the table. These include
Hide/Show: Toggles (hides) the right-hand side Details Pane.
Download: Downloads the selected items (rows).
Copy: Copies the selected items to the clipboard.
Group: Opens a pop-up window to enable adding the selected sequences to an existing or new group in the private workspace.
Genome: Loads the Genome View Overview page corresponding to the selected feature. Available only if a single feature is selected.
Genomes: Loads the Genomes Table, listing the genomes that correspond to the selected features. Available only if multiple features are selected.
More details are available in the Action Buttons user guide.
References¶
Ondov BD, Treangen TJ, Melsted P et al. Mash: fast genome and metagenome distance estimation using MinHash, Genome biology 2016;17:132.